Containers

What are containers
Containerization is a software deployment process that bundles an application's code with all the files and libraries it needs to run on any infrastructure. It makes the application less dependent on the OS and system-wide installed libraries. A particular application packed together with its libraries and other files is a container. An image is a file sitting somewhere on the disk. A container is a runtime instance of an image.
Singularity is an open-source program for containerization used in MetaCentrum.
Singularity is invoked by the command singularity.
Singularity images (= containers) are commonly suffixed by .sif.
Singularity can import Docker images without having Docker installed or being a superuser
- see the Docker part.
Direct Singularity usage (new feature)
Singularity image can be added as PBS parameter
The Singularity image can now be specified directly upon job submission as a PBS parameter, and the container will be automatically launched when the job starts. This is a new feature (Aug 2024). Keep in mind that this new feature is still in testing mode, and probably some expansion and modifications (e.g. towards parallelized computing) will be still under way.
Key features:
- run the job as
qsub -v PBS_SINGULARITY_IMAGE=<singularity_image>,<singularity_image>is the location of image, e.g. /cvmfs/singularity.metacentrum.cz/Metacentrum/debian11-openpbs.sifdocker://ubuntu:latest- you can also pass the argument inside batch script as
#PBS -v PBS_SINGULARITY_IMAGE=<singularity_image> /var/spool/pbsand/etc/pbs.confare binded into the container automatically
Example
# I want to run Deb 11 OS from a container
# Currently, the MetaCentrum OS is Deb 12
(BOOKWORM)user_123@tarkil:~$ lsb_release -a
No LSB modules are available.
Distributor ID: Debian
Description: Debian GNU/Linux 12 (bookworm)
Release: 12
Codename: bookworm
# run the job
(BOOKWORM)user_123@tarkil:~$ qsub -I -l select=1:ncpus=1 -l walltime=1:00:00 -v PBS_SINGULARITY_IMAGE=/cvmfs/singularity.metacentrum.cz/Metacentrum/debian11-openpbs.sif
qsub: waiting for job 3696027.pbs-m1.metacentrum.cz to start
qsub: job 3696027.pbs-m1.metacentrum.cz ready
Singularity> lsb_release -a
No LSB modules are available.
Distributor ID: Debian
Description: Debian GNU/Linux 11 (bullseye)
Release: 11
Codename: bullseye
Singularity>
Singularity usage (old way)
In the basic usecases of Singularity images covered below we suppose there already exists an image my_image.sif we intend to use.
Singularity environment variables
Singularity employs a few paths to store various temporary files:
CACHEDIR:- downloaded layersLOCALCACHEDIR: - run shell execTMPDIR: - squashfs and temporary files, there is limit 1GB by default, if you need more use scratch
Tip
If SCRATCHDIR exists, Singularity automatically exports their variables SINGULARITY_TMPDIR and SINGULARITY_CACHEDIR. That is why in the rest of this section the explicit export of these variables is not done.
List options
To list all commands, run
singularity
or
singularity --help
on the command line.
Run a command in the container
Passing a command to a Singularity image is done by singularity exec "command".
For example,
(BULLSEYE)user123@skirit:~$ singularity exec my_image.sif bash -c "java -version"
java version "1.8.0_60"
Open shell in container
You can also open a shell within a container to work interactively. This is done by singularity shell command.
For example,
(BULLSEYE)user123@skirit:~$ singularity shell my_image.sif
Singularity> command_1
Singularity> command_2
...
Singularity> command_N
Do not use frontends for extensive containers' usage
Apart from light testing and learning, running containers right on frontends is equivalent to computing on frontend. This is strongly discouraged. For an extensive work with containers, use interactive or batch job.
Use container in an interactive job
First run interactive job with scratch directory:
qsub -l select=1:scratch_local=10gb -l walltime=24:00:00
What is the scratch directory good for?
You may need to redirect some Singularity environment variables to store temporary files. To point them to SCRATCHDIR is one option. Alternatively you may redirect the Singularity variables to some folder in your home.
Run the container and open a shell within the container for interactive work:
user123@node123:~$ singularity shell my_image.sif
Singularity> command_1
Singularity> command_2
...
Singularity> command_N
Use container in bash job
As with any other software, it is possible to pass a batch script to a container, too.
Assume the batch script resides in /storage/city_N/home/user123/script.sh.
Is the batch script for the Singularity application any different from the script for "normal" application?
In general, no, though for a very simple script, this may be the case. Most often, you will at least need to redirect Singularity environment variables to SCRATCHDIR. The --bind and bash -c commands shown below may be hidden in the script, too.
Variant A: put Singularity-specific option on command line
qsub -l select=1:scratch_local=10GB -l walltime=24:00:00 -- \
export LOCALCACHEDIR=$SCRATCHDIR \
singularity exec -B /storage/city_N/home/user123/script.sh:/home/user123/script.sh \
my_image.sif bash -c "/home/user123/script.sh"
Are you familiar with how the -B (or --bind) option in Singularity works?
The -B option binds the location of your script in the grid directory onto /home/user123/ container directory.
The -B option binds the location of your script in the grid directory onto /home/user123/ container directory.
The container sees itself as a small enclosed operating system with libraries and traditional Linux directories like /home, /tmp or /mnt. However this causes problem if the "real" filesystem is more complex and contains custom folders, like /storage/city_N/home/user123. In this case you have to specifically tell Singularity to bind these un-traditional directories.
The -B option works like "real" directory location:location within container.
Even better, -B /home/user123:/storage/city_N/home/user123 does this:
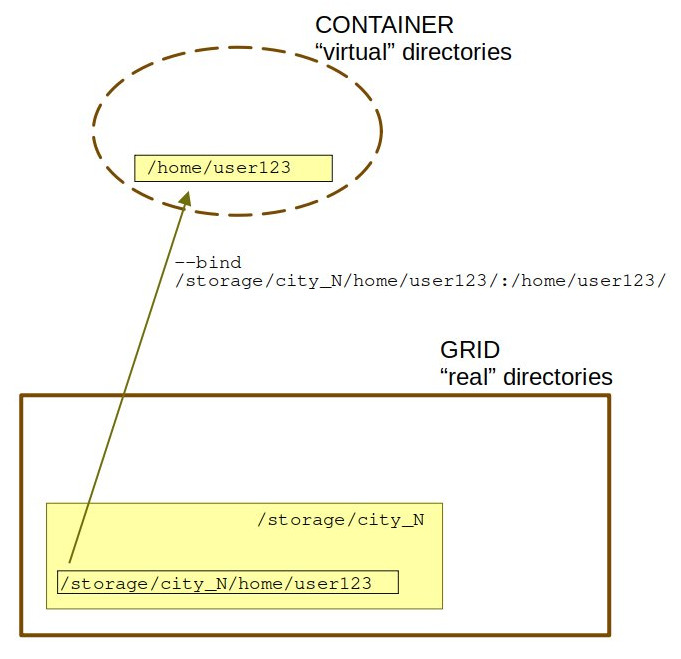
Without the -B option, the container will automatically bind to itself host directories on the computational node where the job is run and the script may not be found.
Variant B: hide Singularity-specific option into the script
Assume the batch script resides in /storage/city_N/home/user123/script.sh AND that you want to have the CLI command as simple as possible.
In this case, run the batch job as
qsub /storage/city_N/home/user123/script.sh
The script /storage/city_N/home/user123/script.sh will then look somehow like the following:
#!/bin/bash
#PBS -N Singularity_Job
#PBS -l select=1:scratch_local=10gb
#PBS -l walltime=24:00:00
# define variables
SING_IMAGE="/path/to/my_image.sif/"
HOMEDIR=/storage/city_N/home/$USER # substitute username and path to to your real username and path
...
# test if scratch directory is set
# if scratch directory is not set, issue error message and exit
test -n "$SCRATCHDIR" || { echo >&2 "Variable SCRATCHDIR is not set!"; exit 1; }
#set SINGULARITY variables for runtime data
export SINGULARITY_CACHEDIR=$HOMEDIR
export SINGULARITY_LOCALCACHEDIR=$SCRATCHDIR
...
singularity exec --bind /storage/ \
$SING_IMAGE/my_image.sif [commands]"
...
Pre-built Singularity images
MetaCentrum offers a couple of ready-to-use Singularity images (.sif files) together with batch scripts to run them.
These images are placed /cvmfs/singularity.metacentrum.cz/.
/cvmfs/singularity.metacentrum.cz/
│
├── BioBakery
│
├── Bioconductor
│
├── Biomex
│
├── BioNano
│
├── BIOP-Desktop
│
├── Braker
│
├── FEFLOW/
| # FEFLOW is a computer program for simulating groundwater flow,
| # mass transfer and heat transfer in porous and fractured media
│
├── GATE
│
├── HybPiper
│
├── KKY/
| # KKY directory contains tools prepared specifically for
| # Department of Cybernetics (KKY) at University of West Bohemia in Pilsen
│
├── Metacentrum
| # OSs
│
├── NGC/
| # NGC are GPU-tuned NVIDIA frameworks for deep learning
| # packed as containers,
| # including NAMD3, OpenCV, PyTorch, qEspresso, TensorFlow
│
├── NVIDIA_HPC_SDK/
| # an NVIDIA comprehensive suite of compilers, libraries and tools for HPC
│
├── OpenFOAM/
| # OpenFOAM is a software for modelling complex fluid flows,
| # chemical reactions, turbulence and heat transfer,
| # solid dynamics, electromagnetics and more.
│
├── Peregrine/
| # Peregrine is a fast genome assembler for accurate long reads
│
├── RepeatExplorer
│
├── RNApeg/
| # RNApeg is an RNA junction calling, correction, and quality-control package
|
├── Rosetta
│
├── RoseTTAFold2
│
├── RStudio/
| # Integrated development environment (IDE) for R
│
├── TE-Tools/
| # Dfam TE Tools is a container
| # that includes RepeatMasker, RepeatModeler, and coseg
│
├── Trinity
Docker usage
Docker is just another type of container designed for single-user system that requires root priviliges.
For safety reasons, running Docker images directly is not possible on Metacentrum nodes.
If you need to use a Docker image in MetaCentrum, either
- run it in Singularity as described below, or
- use Kubernetes service.
Kubernetes service
Kubernetes is a Docker-specialized platform based on Kubernetes orchestrator.
In general, the platform can be used for almost anything, starting with a simple web application, to running a full remote desktop or a complex workflow pipeline.
Run in Singularity
If you have an existing Docker image, you can run it without root privileges in Singularity.
If you want to change something or make your own image from scratch, you'll need root privileges; go to Kubernetes.
Prepare Singularity image from Docker image as:
singularity build image.img docker://tensorflow/tensorflow:latest
Start docker image as:
qsub -l select=1 -l walltime=24:00:00 -- /usr/bin/singularity exec docker://ubuntu:latest echo "Hello Dinosaur!"
Docker download instructions of the type
docker pull sangerpathogens/circlator
are in Singularity replaced as
singularity pull docker://sangerpathogens/circlator
This command will create circlator_latest.sif, a singularity image of docker image.
Docker mounting command of the type
docker run -v /home/ubuntu/data:/data sangerpathogens/circlator
are in Singularity replaced by
mkdir circ_read; singularity run -B ./circ_read/:/data ./circlator_latest.sif
where circ_read is the folder used for getting data into an image. By running the command you are in the image and using df -h you can check that the folder is mounted.
For more details see https://www.sylabs.io/guides/3.7/user-guide/singularity_and_docker.html.
Example
Let's say you want to run Chronusq tool, an quantum chemical software package.
This is a small tool and can be compiled directly on the frontend. The resulting .sif image will be about 166 MB large.
Alternatively, you can setup and interactive job and work from within the job.
First set path for temporary files:
(Default is /tmp which has quota of only 1 GB.)
export SINGULARITY_TMPDIR=/storage/CITY_XY/home/user123/
If you work within interactive job with scratch directory, then export SINGULARITY_TMPDIR=$SCRATCHDIR is done automatically.
Next, download Docker image and build
singularity pull docker://uwligroup/chronusq
After ca 5 minus the singularity image chronusq_latest.sif will be ready.
To test the image, download water molecule water.inp from here or copy from here:
[Molecule]
charge = 0
mult = 1
geom:
O 0.0 -0.076 0.0
H 0.867 0.601 0.0
H -0.867 0.601 0.0
[QM]
reference = Real RHF
job = SCF
[BASIS]
basis = 6-31G(d)
[MISC]
nsmp = 1
mem = 100 MB
and run the calculation of single-point ground state energy as
singularity exec chronusq_latest.sif chronusq water.inp
Advanced options
Custom Singularity build
Preparing your own Singularity image is intended for experienced users. Reading Singularity documentation is a good idea too.
To build custom image, root privileges are needed, but this can be replaced by User Namespace Remapping. For this purpose MetaCentrum offers a server builder.metacentrum.cz with User Namespace Remapping.
Warning
Access to builder.metacentrum.cz is not granted automatically. Users intending to build Singularity images have to apply for membership in group builders by writing a request to meta@cesnet.cz.
Image file is read-only and to modify it you have to use sandbox directory. In this example we use Debian Buster docker image.
singularity build -f -s test1.sbox docker://debian:buster
After running container from sandbox, you can make changes via package system, compile the source codes, install pip/conda modules etc. In this example we install ffmpeg package in Debian Buster environment.
It is necessary to work in the scratch directory, using fakeroot mode (-f options) is not possible on storages (/storage/cityXY/...).
singularity build -f -s test1.sbox docker://debian:buster
After running container from sandbox, you can make changes via package system, compile the source codes, install pip/conda modules etc. In this example we install ffmpeg package in Debian Buster environment.
singularity shell -f -w test1.sbox
Singularity> apt update && apt install ffmpeg
Singularity> exit
Build image from sandbox:
singularity build -f test1.SIF test1.sbox
For the repeatable build of image you can use the definition file, example test1.def
Bootstrap: docker
From: debian:buster
%post
apt-get update && apt-get install -y ffmpeg
Build image from recipe file:
singularity build –f test1.SIF test1.def
For more details, see https://sylabs.io/guides/3.7/user-guide/definition_files.html.
Singularity parallelized job
The scenario for this setup is: two nodes with common scratch dir
#!/bin/bash
#PBS -l select=2:ncpus=2:mem=1gb:scratch_shared=4gb
#PBS -l walltime=04:00:00
#PBS -l place=scatter
# modify/delete the above given guidelines according to your job's needs
module add openmpi/2.0.1-gcc
cat $PBS_NODEFILE |uniq >nodes.txt
# run job over ethernet or infiniband (mpirun autoselects better)
mpirun -n 2 --hostfile nodes.txt singularity exec my_image.img /path/to/program