AlphaFold
ls /storage/brno11-elixir/projects/alphafold/ # available as Singularity imageAlphaFold is a cutting-edge software tool designed to predict 3D structures of proteins. Developed by DeepMind, AlphaFold has revolutionized the field of structural biology by providing accurate predictions of 3D models of protein structures, which is crucial for understanding their functions and interactions.
Alphafold in Kubernetes
As as a first choice, we recommend to run Alphafold via Kubernetes web application Foldify.
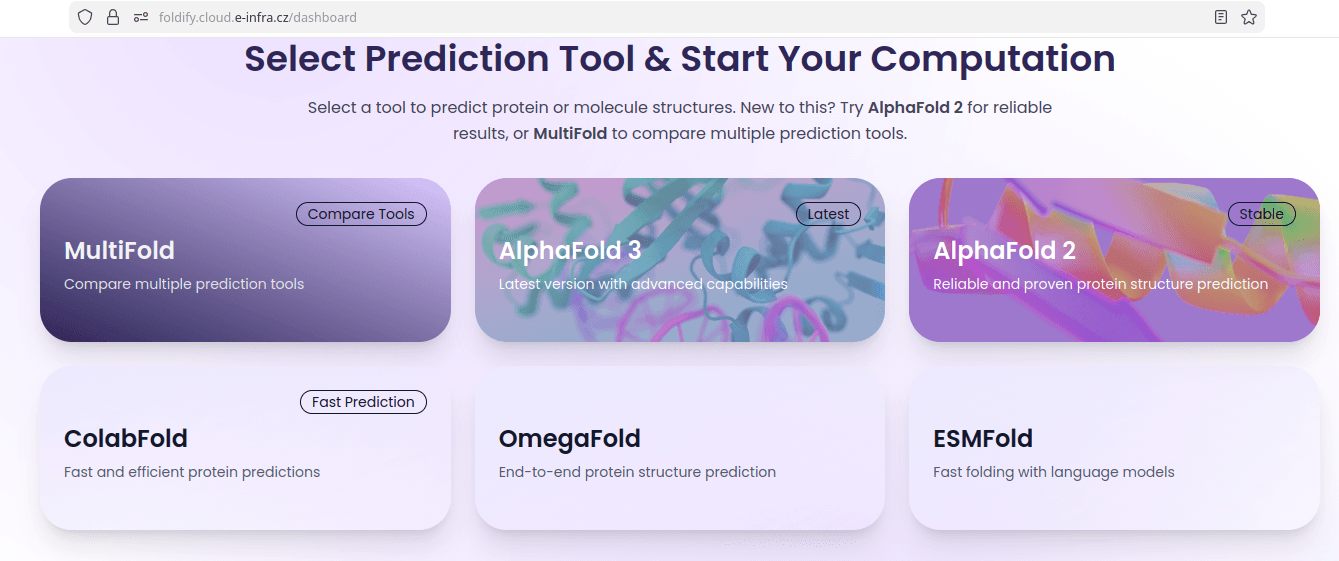
Foldify provides a suite of tools, including:
- AlphaFold3
- AlphaFold2
- ColabFold
- OmegaFold
- ESMFold
as well as integrated tool to compare between models (MultiFold).
Alphafold in OnDemand
Another option is to run Alphafold on the grid infrastructure via OnDemand service.
Follow these steps:
- Access the OnDemand dashboard and navigate to the
Job Composer->New job->From template.
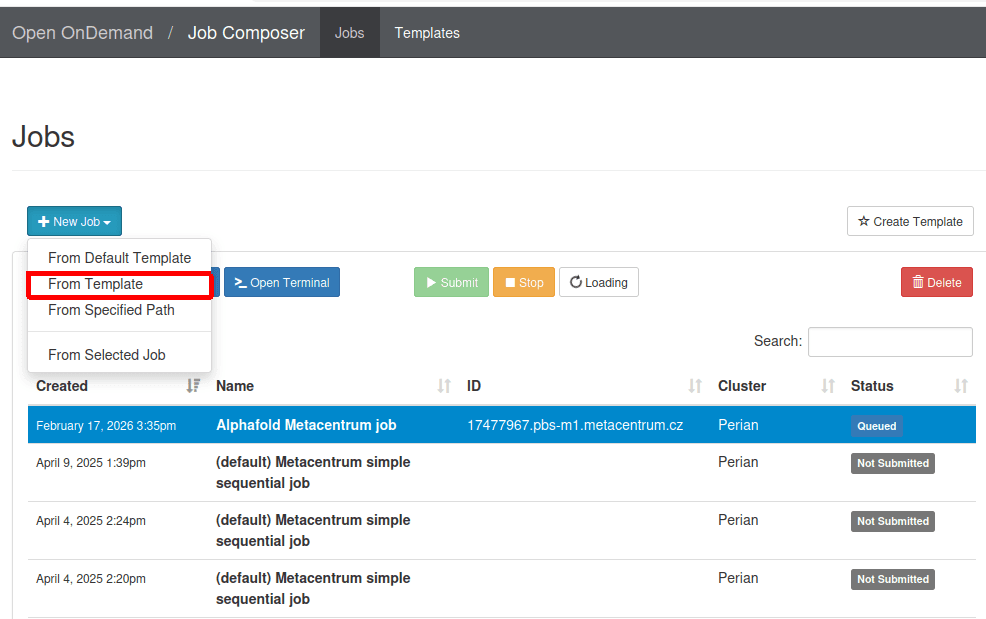
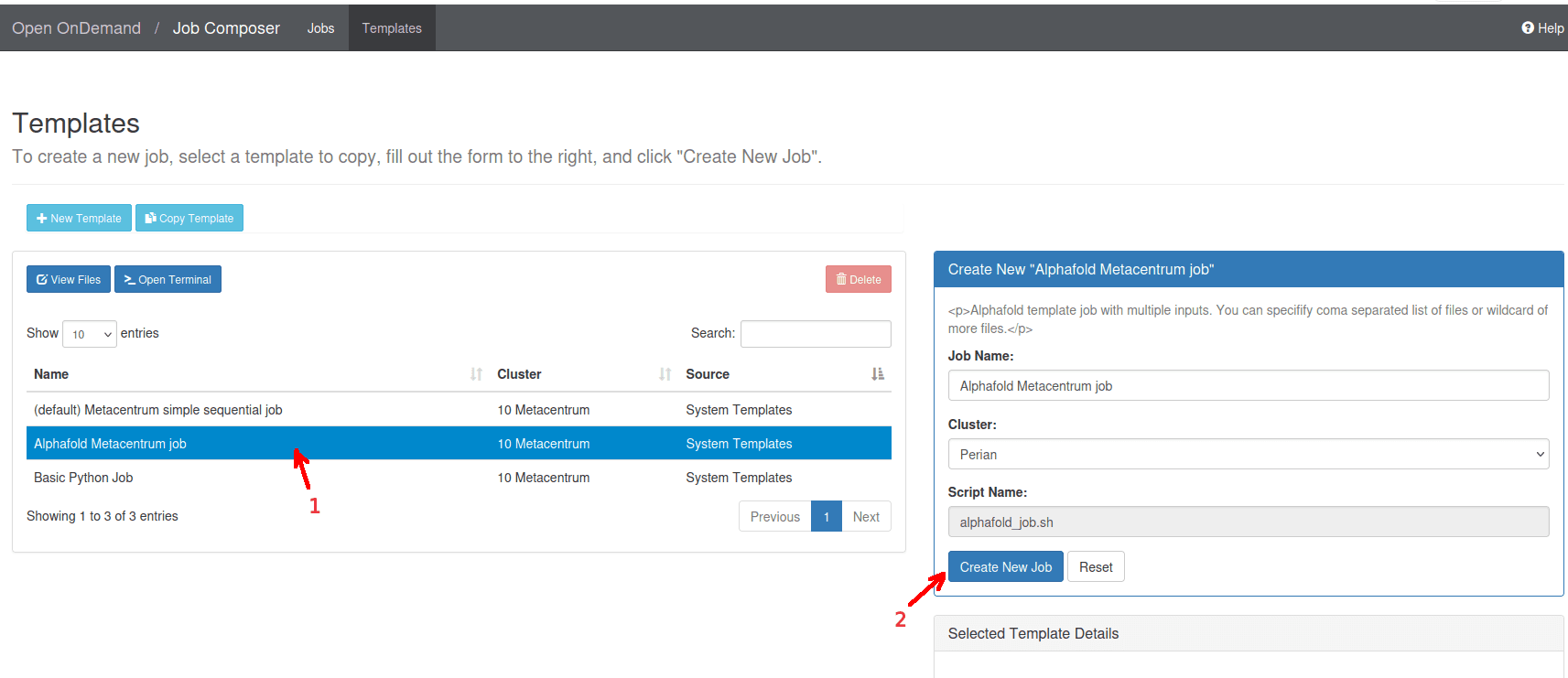
- From a list of templates, choose
Alphafold MetaCentrum job->Create New Job.
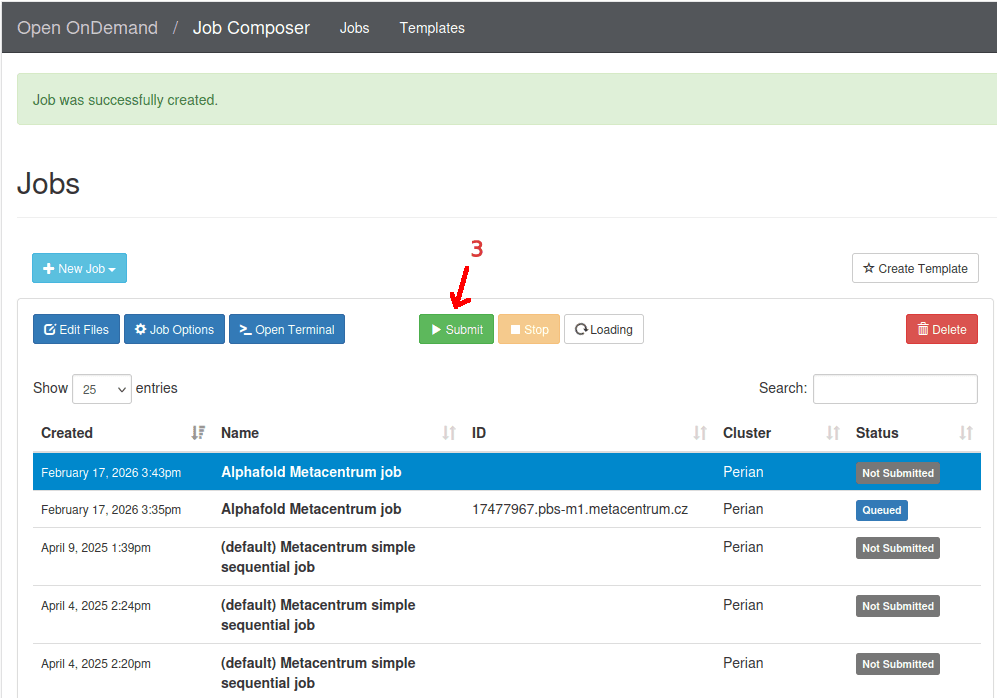
- Finally, select the newly created job on the top of a list and click
Submit..
Alphafold as Singularity image in Remote desktop
The .sif images of Alphafor reside in /storage/brno11-elixir/projects/alphafold directory together with databases and example scripts . This is where the OnDemand takes the images from.
Using Remote desktop, AlphaFold can be also run from a Singularity image manually. This option is recommended to advanced users or as a fallback option in case OnDemand and/or Kubernetes service are down.
CUDA license
To use Alphafold with CUDA, you will also need to accept license for cuDNN library. You will find the link on MetaVO licence pages.
Models
You can control which AlphaFold model to run by adding the --model_preset= flag. We provide the following models:
monomer: This is the original model used at CASP14 with no ensembling.monomer_casp14: This is the original model used at CASP14 withnum_ensemble=8, matching our CASP14 configuration. This is largely provided for reproducibility as it is 8x more computationally expensive for limited accuracy gain (+0.1 average GDT gain on CASP14 domains).monomer_ptm: This is the original CASP14 model fine tuned with the pTM head, providing a pairwise confidence measure. It is slightly less accurate than the normal monomer model.multimer: This is the AlphaFold-Multimer model. To use this model, provide a multi-sequence FASTA file. In addition, the UniProt database should have been downloaded.
Speed/Quality
You can control MSA speed/quality tradeoff by adding --db_preset=reduced_dbs or --db_preset=full_dbs to the run command. We provide the following presets:
reduced_dbs: This preset is optimized for speed and lower hardware requirements. It runs with a reduced version of the BFD database. It requires 8 CPU cores (vCPUs), 8 GB of RAM, and 600 GB of disk space.full_dbs: This runs with all genetic databases used at CASP14.
Comparison between models
Example of run with different models and speed/quality tradeoff with example file seq.fasta, multimer with multi.fasta. This test shows difference of RAM consuming a length of run with the same test.
| model | speed/quality | RAM | Duration | cluster - GPU |
|---|---|---|---|---|
| monomer | full_dbs | 185 GB | 28 min | glados - RTX2080 - 8GB |
| monomer | reduced_dbs | 153 GB | 36 min | glados - RTX2080 - 8GB |
| monomer_casp14 | full_dbs | 197 GB | 35 min | zia - A100 - 40GB |
| monomer_casp14 | reduced_dbs | 38 GB | 36 min | zia - A100 - 40GB |
| monomer_ptm | full_dbs | 190 GB | 36 min | gita - RTX2080 Ti - 11GB |
| monomer_ptm | reduced_dbs | 55 GB | 32 min | gita - RTX2080 Ti - 11GB |
| multimer | full_dbs | 119 GB | 75 min | zia - A100 - 40GB |
| multimer | reduced_dbs | 40 GB | 73 min | zia - A100 - 40GB |
How to speedup the JACKHMMR step
The JACKHMMER module quite old and not optimized for current machines, so it is rarely able to utilize more than 3 CPUs, thus making it difficult for Alphafold to use provided CPUs efficiently. Alphafold increases parallelism by running jackhmmer invocations for various databases concurrently, but it is not perfect since the databases are of different size. Also, there are steps which are not parallel at all, e.g. template search and construction.
There are, however, ways to to improve CPU utilization of your jobs a little:
- Use unpacked
mmcifdatabase
DATABASES_DIR2="/storage/brno11-elixir/projects/alphafold/alphafold.db-3.0.1
...
--db_dir=/public_databases2
--pdb_database_path=/public_databases2/mmcif_files- Avoid JACKHMMR step using unofficial databases
If you do not mind using unofficial databases (related to MMseqs2, not identical to those used by original Alphafold3 with JACKHMMER), there is potential for massive speedup: you can avoid slow JACKHMMER step by preprocessing your input.
Add MSAs from our mmseqs server, the use out.json with AF3:
mamba activate abcfold-1.0.8
python $CONDA_PREFIX/lib/python3.11/site-packages/abcfold/scripts/add_mmseqs_msa.py --input_json in.json --output_json out.json --cerit_serverLast updated on
